Gplots Library
Browse other questions tagged r gplots or ask your own question. The Overflow Blog Podcast 307: Owning the code, from integration to delivery.
- OK, can't help you then sorry. Sometimes folk are confused between install and attach when they're complete beginners so it's worth checking. You're obviously not.
- Gplots: Various R Programming Tools for Plotting Data Various R programming tools for plotting data, including: - calculating and plotting locally smoothed summary.
- Install the R package from Github. Depends: Rsamtools, GenomicFeatures (= 1.14.5), Rcpp, ggplot2, doParallel, foreach, gplots, RColorBrewer, RcppArmadillo.
There are several color palettes available in R such as rainbow(), heat.colors(), terrain.colors(), and topo.colors(). We can visualize these as 3D pie charts using the plotrix R package.
Other popular color palettes include the RColorBrewer package that has a variety of sequential, divergent and qualitative palettes and the wesandersonpackage that has color palettes derived from his films.
You can also create your own color palettes in R with your colors of choice with the colors() function or creating a vector with the color names. A great review and cheat sheet for R colors can be found at http://research.stowers-institute.org/efg/R/Color/Chart/.
You can also create color palettes with hex color codes. A great example of this is to work with popular color palettes available on the http://www.colorcombos.com website. This website has various palettes you can choose from and even derive color palettes from your favorite websites. For example, let’s grab the color palette from the rjbioinformatics.com website at http://www.colorcombos.com/grabcolors.html .
After entering the URL of our website, we will receive the hex codes for the color scheme used on the website.
We can even export the colors as little pencils 🙂

You can also choose from hundred of color schemes based on your color of choice. For example, we will also create a color palette based on the color olive – ColorCombo382.
We can also create a color palette with the colorRampPalette() to use for heatmaps and other plots. For this example, we will use the leukemia dataset available in the GSVAdata package, which corresponds to microarray data from 37 human acute leukemias where 20 of these cases are Acute lymphoblastic leukemia (ALL) and the other 17 are ALL’s with Mixed leukemia gene rearrangements. For more information on the study please see Armstrong et al. Nat Genet 30:41-47, 2002.
Now you are all set to work with and create your own awesome color palettes! Happy R programing 🙂
At this point we have one or more sets of DEGs from our experiment. It might be just one set or it might be a large number of comparisons made with a set of DEGs per comparison and/ or results from multiple approaches (manual, packages). This chapter offers some guidance to visualize, summarize and - finally - connect some biological relevance to the results. Browse this chapter first before choosing the visualization(s) to make for your data! If your article contains relevant figures that you would like to try and reproduce (given that enough data is available) that can be done as well instead of the visualizations shown here. Note however that the Volcano plot described below is required.
5.1 Volcano Plot
A volcano plot is often the first visualization of the data once the statistical tests are completed. This plot shows data for all genes and we highlight those genes that are considered DEG by using thresholds for both the (adjusted) p-value and a fold-change. Many articles describe values used for these thresholds in their methods section, otherwise a good default is 0.05 for the adjusted p-value and around 1.0 for the log-FC.
This scatterplot places the -log10(pvalue) values on the y-axis and the log-FC on the x-axis. Coloring is done based on the thresholds (-log10(0.05) for the pvalue and log2(2) for the log-FC).
The code below shows how to get data from DESeq2 and format this in a usable dataframe to create the figures below. Basically it’s a combination of rlog-normalized count data from the rld-object (to calculate the fold-changes manually, see the heatmap below), results from DESeq2 (to filter for significant genes) and the Entrez IDs that we will use in the pathway analysisfor all genes. The threshold variable is used to subset the complete dataset for DEGs that are also filtered for their log-FC value (stored in the deg dataframe).
Quitting from lines 1169-1171 (gene_expression.Rmd)Error in split(data, ceiling(seq_along(data)/nrow)) :object ‘deseq.degs’ not foundCalls: … withVisible -> eval -> eval -> pander -> vector.to.table -> splitIn addition: Warning messages:1: In yaml.load(readLines(con), error.label = error.label, …) :R expressions in yaml.load will not be auto-evaluated by default in the near future2: In yaml.load(readLines(con), error.label = error.label, …) :R expressions in yaml.load will not be auto-evaluated by default in the near future
The volcano plot below is created on the complete dataset (all genes from the deseq.results dataframe from the chunk above) where the coloring is done based on the threshold variable. Genes with an adjusted p-value of 1 (shown as log10(1)) are included (shown at y=0). It is perfectly fine to use the standard R plot method combined with the abline function to separate the DEGs from the other genes. If you already have experience with ggplot2, you can use the code below to reproduce this image, otherwise, there is a vanilla R solution further below.
Notes:* When you have used edgeR instead you need to change the names of the columns (i.e. log2FoldChangelogFC and padjFDR) used in this example.* In this example there are too many DEGs to annotate with their name in the plot. Therefore the last line (call to the geom_text_repel function) uses a more strict selection to limit the number of labels, just for demonstration purposes. Disable labels completely if you have > 50 DEGs determined by both the log fold change and adjusted p-value or only label those genes showing the biggest/ most significant effect* Also, this example limits the ranges on both the x- and y-axis (xlim and ylim functions) which might hide data, do not use this setting in your final report.
Figure 5.1: ggplot2 Volcano plot for DESeq2 output
As an alternative method that uses standard R plotting with color-coding the DEGs can be done by adding points for each group separately and giving them a certain color. By using the with function we don’t have to type deseq.results$padj but can suffice with typing padj to access this column. The first plot command plots all the points in black and we overwrite points in different colors with the points function later on.
Figure 5.2: Vanilla R Volcano plot for DESeq2 output
5.2 Venn Diagram
Another common visualization is a Venn-diagram. In this document for instance, both DESeq2 and edgeR have been used to find DEGs. To quickly compare the results from these packages we can create a single diagram showing how many DEGs are found by both packages and - also interesting - the number of genes (amount, not which) that are uniquely found by both approaches. Most likely you will be comparing DEGs between groups instead of methods. For our example dataset, this could have been comparing DEGs comparing IPSC trisomic vs IPSC disomic DEGs with NEUR trisomic vs NEUR disomic DEGs.
Creating a Venn-diagram can be a good method for selecting a (sub)set of DEGs, for instance by selecting the set of DEGs that are shared between groups (the Intersection, see the Wikipedia page on Set Theory or the other way around, only the DEGs that are unique for a group (the Difference). Keep this in mind when continuing on to the clustering section below where it is most likely not useful to cluster all combinations you might have.
Code examples:* Data preparation (first code-chunk): creating vectors of DEGs for both packages. These DEGs are the gene names (taken from the rownames of the edgeR and DESeq2 outputs)* Using the VennDiagram library (second code-chunk): good for complex/ large number of comparison, highly configurable but more difficult to use. To see some examples, run the following function in the Console: example('draw.quad.venn')* Using the gplots library (third code-chunk): Easier to use and support for more complex Venn diagrams, but not good for adjusting appearance.
The two created vectors of rownames can be used by either taking their lengths (for the VennDiagram library) or just their names (for the gplots library).
Figure 5.3: Venn diagram comparing edgeR and DESeq2 DEGs using ‘VennDiagram’
As mentioned, following is an easier alternative (especially when you have > 2 groups) with the venn function from the gplots library. This only needs a list containing either the row-numbers or the gene-names of the DEGs which is easier (but offers less adjustability to make it prettier).
Figure 5.4: Venn diagram comparing edgeR and DESeq2 DEGs using ‘gplots’
While there is an almost complete overlap between both techniques, DESeq2 finds a total of >700 genes that edgeR does not denote as DEG. Besides the difference in nuber of DEGs, there is also a notable difference in the calculated log fold change values. The following boxplot shows - for all DEGs - the distribution of the log-FC values where DESeq2 shows a far wider range of values. This is possibly due to the large difference in number of DEGs but can also be attributed to different normalization techniques and the many choices of models to choose from for the analysis.
Figure 5.5: Boxplot comparing spread of log fold change values
Let’s quickly compare these log-FC values on a per-gene basis including manually calculated from the DESeq2 rlog and the edgeR cpm methods.
| Gene | edgeR.lFC | DESeq2.lFC | rlog.lFC | cpm.lFC |
|---|---|---|---|---|
| PCDHA10 | -3.602 | -3.631 | -1.985 | -3.527 |
| CHGA | 2.923 | 2.83 | 1.999 | 2.99 |
| ARRB1 | -3.914 | -3.901 | -2.679 | -3.786 |
| TSSC2 | 3.175 | 3.095 | 2.354 | 3.23 |
| LRFN5 | -4.638 | -4.621 | -3.077 | -4.534 |
| C11orf54 | -1.953 | -1.967 | -1.455 | -1.889 |
| MAP2 | 2.647 | 2.552 | 1.253 | 2.708 |
| WNT3 | -3.488 | -3.464 | -2.478 | -3.383 |
| TFAP2C | -2.984 | -2.955 | -2.049 | -2.993 |
| ASAP2 | 1.352 | 1.27 | 0.8311 | 1.412 |
Comparing lFC-values from edgeR, DESeq2 and manually using rlog- and cpm-normalized data
5.3 Clustering
One of the basic visualization steps to show expression data and patterns accross samples. Here, we can create a heatmap of the found DEGs for all samples where the colors show the fold-change value. Usually this heatmap is shown with a two color gradient; from red (downregulated) to green (upregulated). This is however optional and using the default colors from pheatmap for example is perfectly fine as long as a proper legend is present.
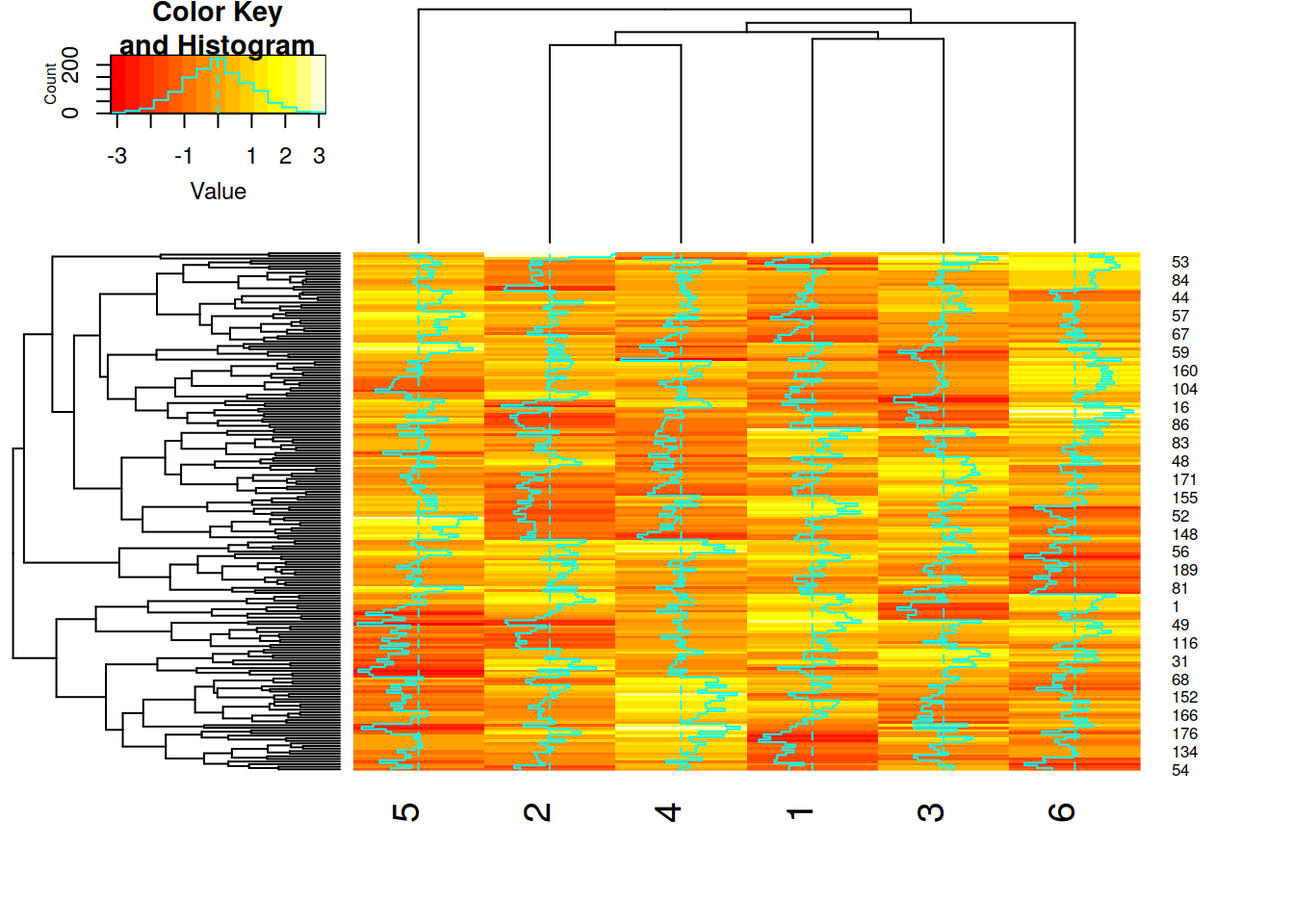
There are multiple methods of creating a heatmap (one of which you’ve already used) and most of these directly apply clustering in the visualization. This clustering can be applied to the expression pattern of a gene (row-clustering), the expression pattern of a sample (column-clustering) or both (default for pheatmap). The first example shown here uses column-based clustering as it shows the replicates for multiple groups (IPSC disomic and trisomic) with their rlog-transformed expression values for the top 30 genes as found by DESeq2. This first example allows an overview of the expression patterns for the DEGs and to see the differences in expression between replicates. It is expected that the replicates for each group cluster together as shown in this example. Genes (rows) are clustered based on their expression values.
Figure 5.6: Heatmap showing the expression values of all disomic and trisomic IPSC samples
This second example is more commonly found in publications as this shows the log fold change values for the comparison that was done. The first three columns shown in this heatmap are the log fold changes of the separate IPSC trisomic replicates vs the average IPSC disomic expression. The 4th column shows the log fold change values for the average IPSC trisomic vs average IPSC disomic samples. Note that column-clustering has been turned off for this type of plot by setting cluster_cols = FALSE (all replicates are from the same group, clustering will not add any useful information).

Figure 5.7: Heatmap showing the Log Fold Changes of all trisomic vs averaged disomic IPSC samples and the average trisomic vs average disomic IPSC samples
A limitation of a heatmap will show itself when more then 100 DEGs are found; this just doesn’t fit well in a single figure and causes the clustering and especially the gene names/ labels to be unreadable. It is therefore always adviced to not only filter on the adjusted p-value, but also on the log-FC value to reduce the number of genes shown. Alternatively if the data is properly annotated, instead of showing gene names for each row, showing a GO-term clustering might reveal expression patterns for certain gene groups.
5.4 Pathway Analysis
In some cases you’ll see hundreds or even thousands of DEGs as a result of an analysis. These large amounts of DEGs are too much for most visualization approaches or to easily say something about the biological context for that many genes.
One approach for tackling such a large set of DEGs is pathway analysis where through different methods genes are grouped by pathway to get an overview of affected pathways in the experiment.
This section will demonstrate two methods for this analysis, one using an online platform for gene-annotation enrichment analysis and an R-method for signaling pathway impact analysis.

5.4.1 Gene Functional Classification using DAVID (>100 DEGs)
The online Database for Annotation, Visualization and Integrated DiscoveryDAVID tool suite allows for multiple - functional - annotation methods, one of these can be used for enrichment analysis based on gene annotation. To use the DAVID tools we need to have the list of gene names, ID’s or other identifyable information (no need for p-values or fold-changes). The basics of this method consist of grouping genes in pathways, count how many genes are in each pathway and compare this with what you would expect if you take a random sample from the genome with the same size as the number of your DEGs.
For example, if the RNA transport pathway contains a total of 50 genes out of the total of 25.000 for the whole mouse genome. This means 0.2% of all mouse genes have a role in this pathway.
If you uploaded a total of 1000 DEGs for analysis, you expect 0.2% = 2 genes from your DEG list to be placed in this pathway. But, it seems that there are 10 genes from the RNA transport pathway in the set of 1000 genes, which is 5 times as much as expected. This difference between expected and observed gene counts per pathway allows to say if a particular pathway is enriched.
DAVID provides a single interface to perform multiple different annotation pipelines, see the table below for available types and their use.
The following example code writes the gene symbols for both analysis approaches to a file for upload on DAVID.
Upload data
Click on the Start Analysis button at the top of the DAVID website. Then, copy the genes (A) or upload the file (B) and select the proper identifier type (OFFICIAL_GENE_SYMBOL in this example). Select the Gene List option in Step 3 and click on the Submit List button in Step 4.
This usually results in a popup that mentions that some of your IDs were found in multiple organisms. Close the popup and select your own organism (Mus musculus in this example) from the Species list. From the 940 uploaded gene-symbols, 881 were found in the mouse which we select followed by clicking on the Select Species button.
Select the source (organism) for the genes
The most useful analysis is the Functional Annotation Chart, selectable from the Start Analysis page after uploading the set of genes. The output table shows many (in this case ~560) chart records in which the genes were grouped. These records orginate from various sources, such as GO-terms, Interpro, KEGG-pathways, SMART, etc. See the image below for a quick explaination of what is displayed in such a chart (mostly just a table..).
An interesting statistic to add to this table is the Fold Enrichment that shows how over- or under-represented this group is. Note that this is not based on gene-expression but only on the deviation of the of the amount of genes provided for a certain group from what is expected given the total number of uploaded gene IDs.
DAVID functional annotation chart - Settings
Now, looking at the article for a random example set with GSE ID GSE80128, one of the conclusions mentions the following on their gene-in-research (Igf2as):
It is suggested that Igf2as play a role in energy metabolism, the cell cycle, histone acetylation and muscle contraction pathways.
With almost a 1000 DEGs grouped in 560 different categories, you can find ‘evidence’ for most conclusions and the difficulty here is finding the truly important pathways. Adviced is to only check the KEGG-pathways listed in the table, sort by the corrected p-value and inspect the pathways that look interesting. For this example, amongst others, the following pathways were listed as significant (p-value < 0.05):
- Metabolic pathways: includes many pathways, 100 genes in total with a 1.6-fold enrichment
- Citrate cycle (TCA): major metabolic pathway, 9 genes with a 5.7-fold enrichment
- Cardiac Muscle contraction: 12 genes with a 3.3-fold enrichment
Clicking on the Term in the table for a KEGG-pathway shows a graphical representation of this pathway indicating the found DEGs (blinking, even), see below.
5.4.2 KEGG-pathway Visualization (>10 DEGs)
Knowing the interesting KEGG-pathway(s) upfront (i.e. it is listed in the article) allows for visualizations applied to that selected pathway. For this we can use the pathviewwebsite or Bioconductor library:
“… It maps and renders a wide variety of biological data on relevant pathway graphs. All users need is to supply their data and specify the target pathway. Pathview automatically downloads the pathway graph data, parses the data file, maps user data to the pathway, and render pathway graph with the mapped data.”
This section demonstrates only the use of the bioconductor package for visualizing not only the DEGs onto pathways, but also the change in expression for each gene.
| hsa00010 | hsa00020 |
|---|---|
| Glycolysis / Gluconeogenesis | Citrate cycle (TCA cycle) |

Table continues below
| hsa00030 | hsa00040 | hsa00051 |
|---|---|---|
| Pentose phosphate pathway | Pentose and glucuronateinterconversions | Fructose and mannosemetabolism |
SYMBOL, GENENAME, ENSEMBL, ENSEMBLPROT, UNIGENE, UNIPROT, ACCNUM, ENSEMBLTRANS, REFSEQ, ENZYME, TAIR, PROSITE and ORF
| log2FoldChange | |
|---|---|
| ABAT | 0.7499 |
| ABCC13 | -3.452 |
| ABHD12B | -3.306 |
| AC002480.2 | -0.1612 |
| AC002480.5 | -5.13 |
| AC005062.2 | -1.949 |
It is fine to use the webtool too for this purpose however it is not notacibly easier to use than the R package.
5.4.3 Signaling Pathway Impact Analysis (>20 DEGs)
To also look at the topology of how genes interact with each other, we can use the SPIA Bioconductor package. This package takes a list of Entrez gene IDs for all DEGs and the complete list of Entrez IDs for all present genes and evaluates - per pathway - if genes are involved and if the pathway is either inhibited or activated.
The code below uses data objects that were not always shown before, see the tables below for their contents.
The input sig_genes named vector contents:
| EntrezID | logFC |
|---|---|
| 18 | 0.7499 |
| 150000 | -3.452 |
| 145447 | -3.306 |
| 43 | -3.055 |
| 51412 | 1.847 |
| 114 | -1.926 |
| 146 | 2.754 |
| 60312 | -1.068 |
| 3899 | 1.061 |
| 116986 | 1.222 |
(subset) of DESeq2 DEGs with their Entrez ID and log2(FC) value
And the all_genes vector or Entrez IDs input (note that this does not contain any expression information, just IDs):
55584, 5010, 6320, 387836, 154790, 7461, 79935, 26047, 85445, 78989 and 57699
To start processing all pathways looking for influenced signal pathway, execute the spia function for the proper organism (Mus Musculus, or mmu in this case):
Install Gplots Library In R
Running this function will print a list of (> 100) pathways it will analyze, so this will take a while:
The resulting data frame is ordered by a p-value and contains the following information:
| Name | ID | pSize | NDE | pNDE | tA |
|---|---|---|---|---|---|
| Alzheimer’s disease | 05010 | 160 | 23 | 2.929e-06 | -0.7695 |
| Parkinson’s disease | 05012 | 113 | 19 | 2.002e-06 | 0 |
| Focal adhesion | 04510 | 196 | 22 | 0.0002214 | 9.707 |
| Glioma | 05214 | 63 | 11 | 0.0002007 | -0.6071 |
| Neurotrophin signaling pathway | 04722 | 120 | 15 | 0.000702 | 2.983 |
Top signaling pathways (continued below)
Plots Library
| pPERT | pG | pGFdr | pGFWER | Status |
|---|---|---|---|---|
| 0.548 | 2.302e-05 | 0.001654 | 0.002693 | Inhibited |
| 1 | 2.828e-05 | 0.001654 | 0.003308 | Inhibited |
| 0.018 | 5.352e-05 | 0.002087 | 0.006262 | Activated |
| 0.863 | 0.001674 | 0.04258 | 0.1958 | Inhibited |
| 0.274 | 0.001838 | 0.04258 | 0.2151 | Activated |
Load Library Gplots
To get an overal picture we can create a plot where each pathway is a point which is placed according to the spia calculated over-representation p-value and pNDE (which is the probability to observe at least nDE genes on the pathway). In this case it is obvious that Alzheimer’s and Parkinson’s pathways with ID’s 05010 and 05012 respectively are the most influenced by this experiment, according to this method.
Plot Library Python
Figure 5.8: SPIA two-way evidence plot showing the most influenced pathways